Whole-genome/exome mapping and variant calling Analysis
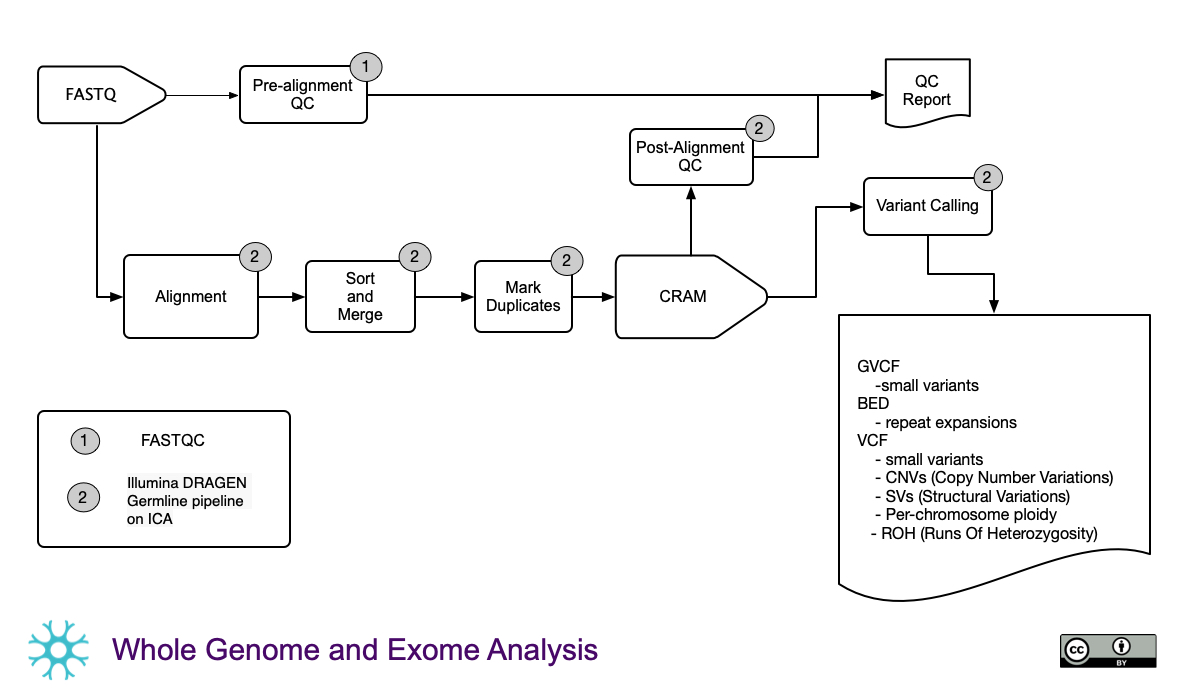
Secondary analysis of WGS DNAseq data using Illumina DRAGEN
Secondary analysis of whole-genome sequencing (WGS) or whole-exome sequencing (WES) is currently commonly performed at the Neuro Bioinformatics Core Facility and requires some input from the users about where the input files are, and what annotations are necessary. Some care should also be given to the control samples you want to use or have available in your own study.
-
Short description of the workflow and its purpose:
-
Run full per-sample secondary analysis pipeline on Illumina DRAGEN Bio-IT, via cloud-based Illumina Connected Analytics (ICA) platform.
- Generates per-sample:
- Alignment
- small variant calls
- Copy-Number Variant (CNV) calls
- Structural Variant (SV) calls
- Repeat expansions
- Per-chromosome ploidy calls
- Runs of heterozygosity
- Various QC metrics
- Raw samples sequences are uploaded to ICA, passed through the Dragen Germline pipeline, then outputs are downloaded back to Compute Canada HPC (beluga server).
Bibliography:
- Original Link where workflow was first presented: Illumina DRAGEN DNA Pipeline
- Reliable variant calling during runtime of Illumina sequencing. Loka TP, Tausch SH, Renard BY. Sci Rep. 2019 Nov 11;9(1):16502 PMID: 31712740 (This publication is not from Illumina, but it represents an early publication that references DRAGEN.)
- From the GATK portal: DRAGEN-GATK – GATK