Single Cell RNA-Seq Analysis
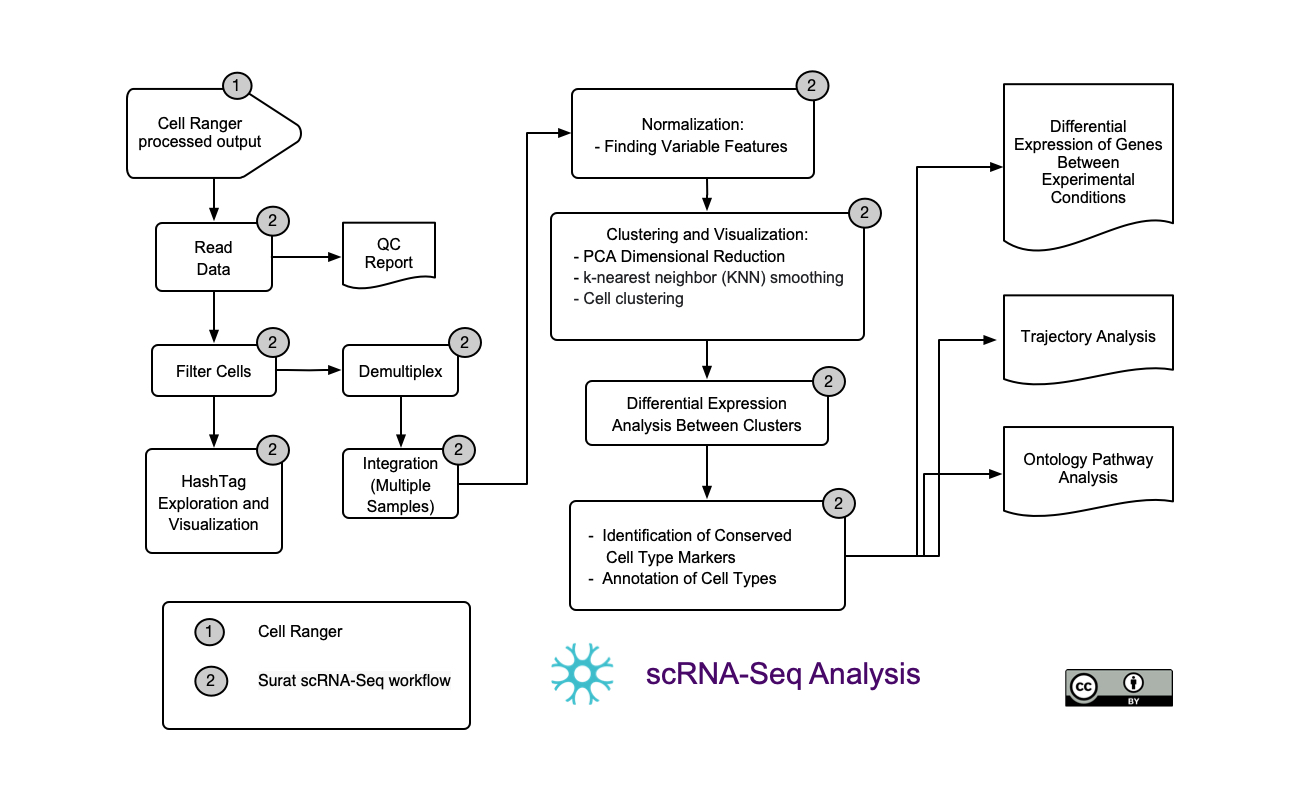
ScRNA-Seq Seurat workflow automation.
Single cell sequencing technologies measure and analyze the molecular signatures in thousands to millions of individual cells. These methods help to characterize the diversity of cell types in simple or complex tissues.
They have been used to study the heterogeneity of brain cell types [1], the immunological system cell diversity [2], or the heterogeneity of cell type composition in different cancer types [3].
Nowadays, ultra-high throughput single cell sequencing technologies can sequence millions of cells or nuclei from a single experiment across different tissues, enabling characterizing the molecular signatures for a wide range of diseases at cellular level [4]. A recently introduced approach, cell Hashing allows uniquely labelling of different experimental samples using oligonucleotide-tagged antibodies to detect cell surface proteins [5]. There exist several bioinformatics tools developed to process and analyze the high-dimensional data generated by sequence alignments and feature counting of each dataset. However, the rising accessibility to this technology and the great variability of applications in medicine and biomedical research creates a need for automatic data processing and analysis.
Here we present a scRNA-seq workflow based on Seurat’s scRNAseq workflow for multiplexed samples [6]. This workflow aims to scale the one dataset analysis to an automated and reproducible analysis for a large number of scRNAseq datasets.
Short description of the workflow and its purpose:
In this workflow we analyze Cell Multiplex data. It takes as input single cell FastQ sequencing files and a feature reference, and it uses Cellranger multi pipeline to produce a matrix of UMI counts per {feature|barcode} pair, in MEX format (raw_feature_bc_matrix files) [7].
This matrix is then the input for the Seurat automation workflow.
The workflow first demultiplexes each sample at a time and performs a quality control (QC) step. Samples demultiplexing is then followed by the integration of all samples data and a downstream integrated analysis. Downstream integrated analysis includes: Dimensional reduction and clustering, cell type identification, DEG, trajectory analysis and ontology pathway analysis.
Tools used in this workflow:
ScRNA-Seq techniques
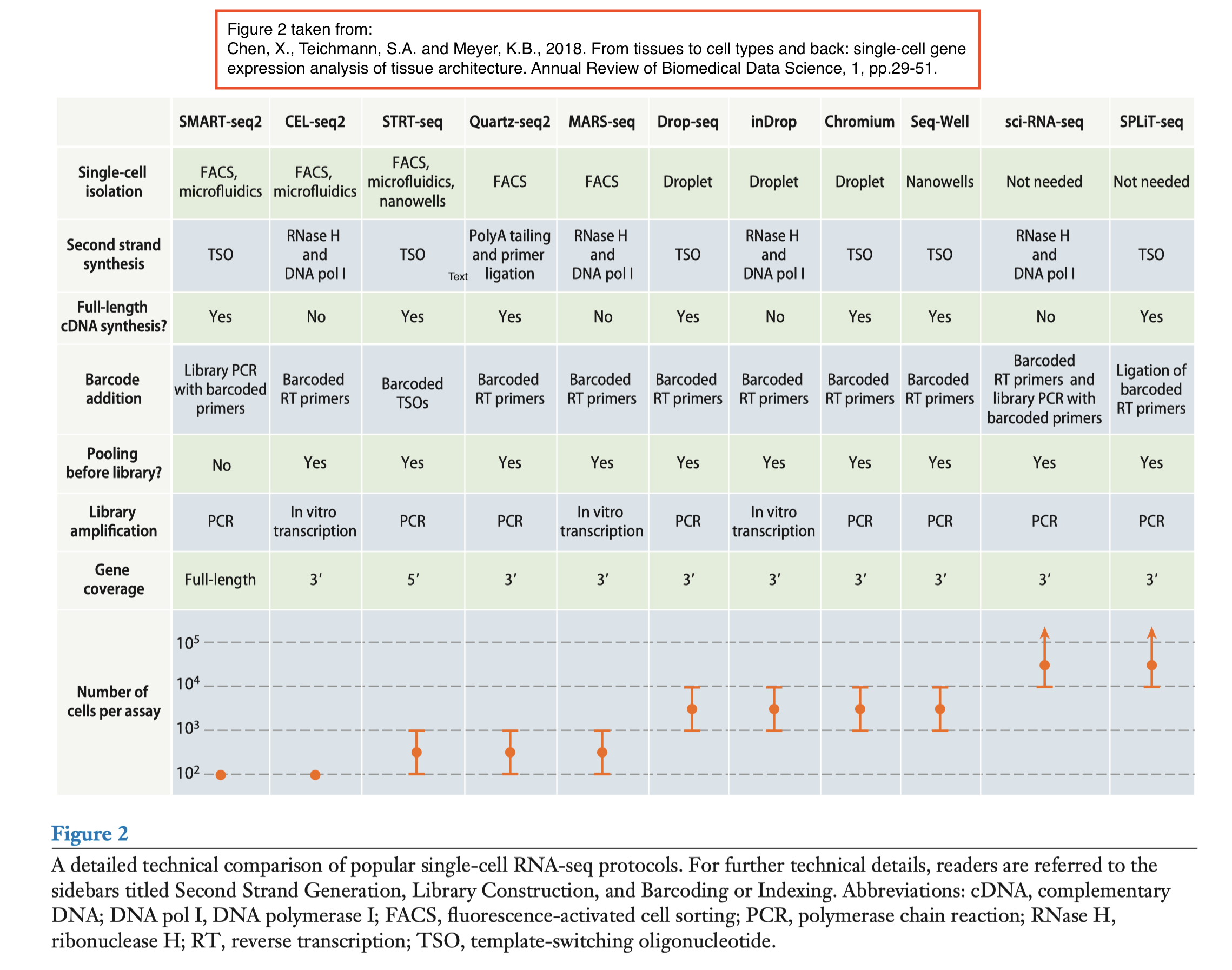
Different techniques are available for generating scRNA data [8][9]:
- CEL-seq2/C1
- Drop-seq
- MARS-seq
- SCRB-seq
- Smart-seq/C1
- Smart-seq2
The difference between these techniques is that:
- Some use unique molecular identifiers (UMIs) versus sequencing full length RNA
- While others are capturing more cells or more genes per cell
- UMIs are a randomly generated barcode added to sequencing libraries before any PCR amplification steps, which enabling the accurate bioinformatic identification of PCR duplicates.
- UMI are a tool for genomic variant detection and by using UMIs instead of actual reads, the output will include less noise and less bias caused by the PCR amplification step.
- Refer to [8] and [9].
Bibliography:
- Armand EJ, Li J, Xie F, Luo C, Mukamel EA. (2021) Single-Cell Sequencing of Brain Cell Transcriptomes and Epigenomes. Neuron. 109, 11–26. PMID: 33412093
- See P, Lum J, Chen J, Ginhoux F. (2018) A Single-Cell Sequencing Guide for Immunologists. Front Immunol [Internet]. 9, 2425–2425. PMID: 30405621
- Wang W, Wang L, She J, Zhu J. (2021) Examining heterogeneity of stromal cells in tumor microenvironment based on pan-cancer single-cell RNA sequencing data. Cancer Biol Med 19, 30–42. Available from: PMID: 34398535
- Ma S-X, Lim SB. (2021) Single-Cell RNA Sequencing in Parkinson’s Disease. Biomedicines. 9. Internet
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, et al. (2018) Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biology. 19, 224. Internet
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. (2015) Spatial reconstruction of single-cell gene expression data. Nature Biotechnology [Internet]. 33, 495–502. Internet
- 10X Genomic. Cell Ranger. What is cellplex. Internet
- Ziegenhain, Christoph, Beate Vieth, Swati Parekh, Björn Reinius, Amy Guillaumet-Adkins, Martha Smets, Heinrich Leonhardt, Holger Heyn, Ines Hellmann, and Wolfgang Enard. “Comparative analysis of single-cell RNA sequencing methods.” Molecular cell 65, no. 4 (2017): 631-643. PMID 28212749
- Chen, X., Teichmann, S.A. and Meyer, K.B., 2018. From tissues to cell types and back: single-cell gene expression analysis of tissue architecture. Annual Review of Biomedical Data Science, 1, pp.29-51. DOI