Split Sequence Analysis
Split-pool ligation-based transcriptome sequencing (SPLiT-seq) is a scRNA-seq method that enables transcriptional profiling of hundreds of thousands of fixed cells or nuclei in a single experiment. SPLiT-seq does not require partitioning single cells into individual compartments (droplets, microwells, or wells) but relies on the cells themselves as compartments , see [1], its steps are presented visually in the following figure [2].
 |
|---|
| split sequence Analysis process |
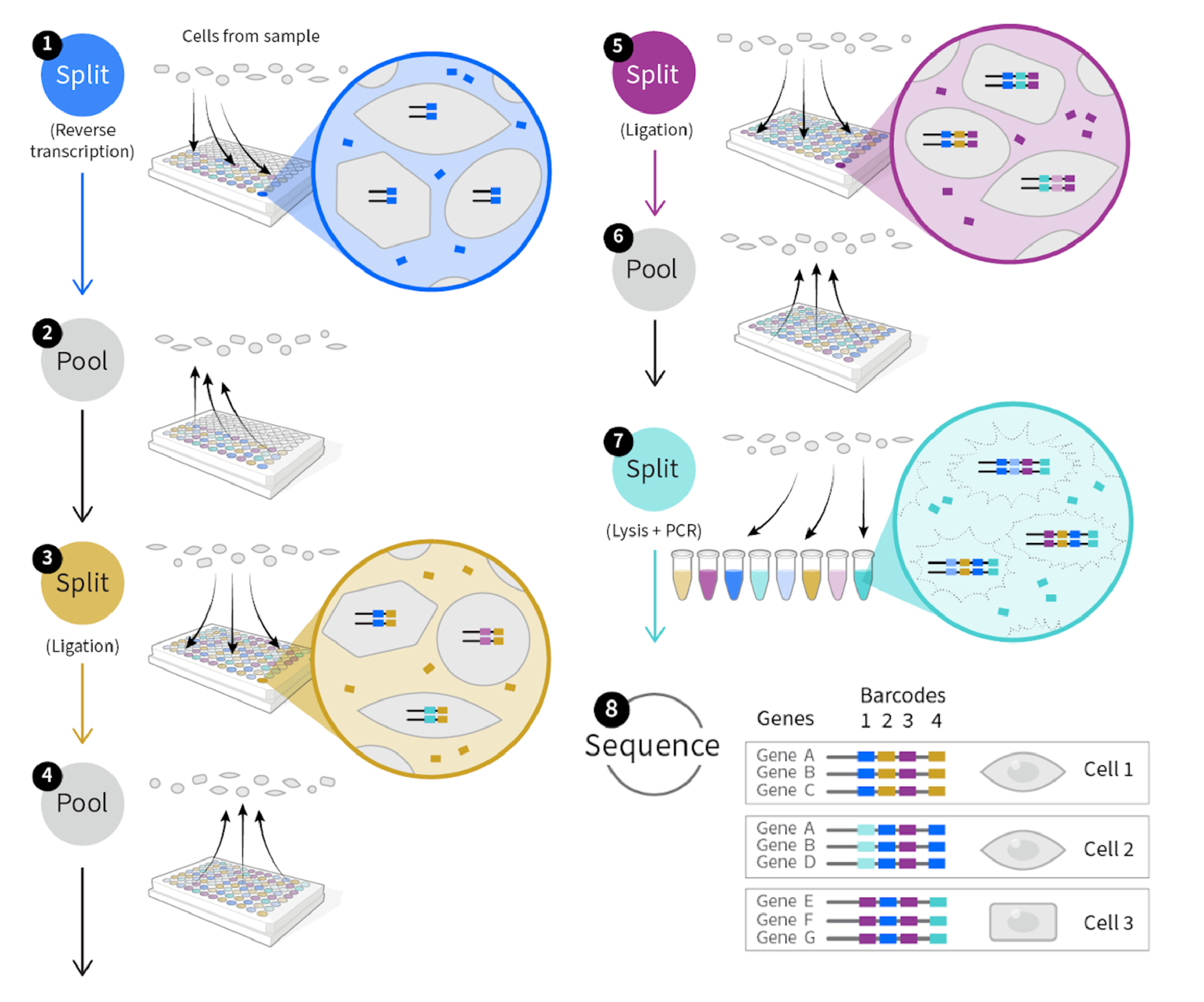
SPLiT-seq has four steps (reference [1]): 1) Reverse transcription: first round of barcoding, cells are distributed into a 96-well plate, and cDNA is generated with an in-cell reverse transcription (RT). 2) Ligation: cells from all wells are pooled and redistributed into a new 96-well plate, where an in-cell ligation reaction appends a second well-specific barcode to the cDNA. 3) Ligation: The third-round barcode, which also contains a unique molecular identifier (UMI), is then appended with another round of pooling, splitt- ing, and ligation. At this step, cells will be pooled and subsequently split into between one and eight distinct populations we call sublibraries. Each sublibrary will contain a different 4th barcode (the Illumina index) and can be sequenced separately, see [3]. 4) Lys and PCR: After three rounds of barcoding, the cells are pooled and split into sublibraries, and sequencing barcodes are introduced by polymerase chain reaction (PCR). This final step pro- vides a fourth barcode, while also making it possible to sequence different numbers of cells in each sublibrary. After sequencing, each transcriptome is assembled by combining reads containing the same four-barcode combination
Analysis Flowchart
![Graphic explanation of the Split-Seq method, [2] scRNA-Seq Analysis](/wf/WF07_split-seq.png) |
|---|
| split sequence Analysis process |
Here we present a split-seq workflow analysis.
Fastq: one use bcl2fastq to generate fastq.-
Prepare reference: run thesplit-pipewith--mode mkrefto create an indexed reference genome for the genome (Homo_sapiens.GRCh38.93.gtf.gz), genes (Homo_sapiens.GRCh38.93.gtf.gz), and a specific version version ofstarandsamtools. Process analyis: in this step you need to run split-pipe to run the following sub-stepspre: Pre-process fastq files (barcode correction + cDNA).align: Align barcode-corrected reads (STAR).post: Post-process read alignment (sort bam, samtools).mol: Molecule info extraction (barcodes, tscps, genes from reads).dge: Digital gene expression matrix creation and stats.ana: Analysis of called cells and samples (clustering, etc).rep: Generate output reports for each sample.clean: Clean up intermediate files (compress / delete).
Thesplit-pipewith--mode all, run all these sub-steps.
Combine: in this step, one needs to combine the results from the processed sublibraries.Analysis output: the output of running the pipeline includes<sample-name>\_analysis_summary.htmlcontains summary statistics and plots for a given sample.<sample-name>\_analysis_summary.csvcontains pipeline metrics about the sample.DGE.mtxis a sparse matrix with cell-gene counts. Each row corresponds to a cell and each column corresponds to a gene (seegenes.csvfile for names/gene-id of each column).genes.csvcontains the gene name, gene id, and genome for each column inDGE.mtx.cell_metadata.csvfile contains information about each cell including the cell barcode, species, sample, well in each round of barcoding, and number of transcript/genes detected.
Tools used in this workflow:
Bibliography:
[1]. Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018 Apr 13;360(6385):176-82.PMID: 29545511
[2]. Single-Cell RNA Sequencing May Be Split By Parse Biosciences Internet
[3]. Parse Biosciences company Internet